The etiology of paroxysmal nocturnal hemoglobinuria (PNH) is somatic PIGA mutations in hematopoietic stem and progenitor cells (HSPCs), and affected PNH-type cells lack cell surface expression of the glycosylphosphatidylinositol-anchored proteins (GPI-APs). In PNH, GPI-AP deficient cells clonally expand and occupy a large proportion of mature cells. Several hypotheses have been proposed to explain clonal expansion of PNH-type HSPCs, including reduced sensitivity to apoptosis and escape from immune-mediated cytotoxicity. However, definitive mechanisms remain unclear. To further understand PNH pathophysiology at high multi-dimensional resolution, we performed single-cell RNA sequencing (scRNA-seq) of bone marrow (BM) cells from PNH patients.
Fresh BM samples were obtained from eight PNH patients (male, n = 2, female, n = 6; median age, 51 [range, 21-67] years), who had large PNH-type granulocyte populations and stable clone sizes (median, 45 [range, 16-99] %), and five age- and sex-matched healthy donors (HDs). PNH-type and wild-type (WT) lineage-CD34+HSPCs were enriched from bone marrow mononuclear cells (BMMNCs) by flow cytometric sorting. Sorted cells were processed through the Chromium Controller (10x genomics, Pleasanton, CA, USA). scRNA-Seq libraries were constructed using the Chromium Single Cell 3' Reagent Kits (10x genomics) according to the manufacturer's instructions. The constructed libraries were sequenced with the NovaSeq 6000 system (Illumina, San Diego, CA, USA).
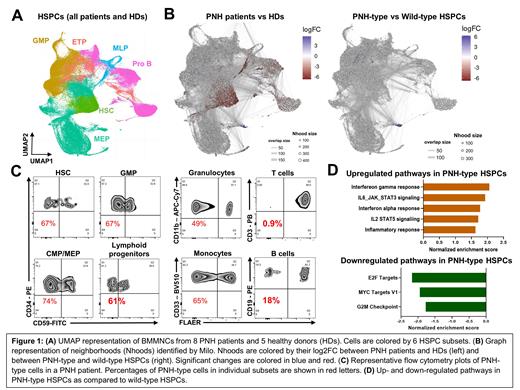
We obtained 81,470 high-quality cells from PNH patients and 56,545 cells from HDs for further analysis. Cells were clustered with Seurat and visualized in UMAP. Cell clusters were assigned to 6 HSPC subsets based on expression of canonical marker genes (Figure 1A). To investigate compositional changes in BMMNCs between PNH patients and HDs, we performed differential abundance analysis with Milo and identified a prominent decrease in hematopoietic stem cells (HSCs) and lymphoid progenitors, as compared to HDs (Figure 1B). We then evaluated differential abundance between PNH-type and WT HSPCs in PNH patients, identifying no significant differences in each HSPC subsets (Figure 1B). These data were consistent with flow cytometric profiling of BMMNCs, similar proportions of PNH-type cells in each HSPC subsets in individual patients despite with profound decrease in PNH-type lymphocytes (Figure 1C).
We next sought to investigate skewed gene sets in PNH-type HSPCs using gene set enrichment analysis. Comparison between PNH-type and WT HSPCs identified upregulation of inflammatory signaling pathways and downregulation of the cell-cycling pathways in PNH-type HSPCs, as compared to WT HSPCs (Figure 1D). We made comparisons within individual subsets. In HSCs, genes involved in IFN-γ response and oxidative phosphorylation were upregulated in PNH-type HSCs, while cell-cycling genes (ie. E2F targets and G2M checkpoint) were not dysregulated, as compared to WT HSCs.
In summary,no significant skewing in lineage differentiation of PNH-type HSPCs was observed, suggesting PNH-type lymphocytes were negatively selected during the maturation process in lymphoid tissues. Upregulation of the inflammatory and immune response pathways in PNH-type HSPCs indicated a proinflammatory phenotype of PNH-type HSPCs. Upregulation of cell cycle-related pathways was not observed in PNH-type HSCs compared to WT HSCs, consistent with stable PNH clone sizes in our cohort of PNH patients.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal